Tutorials
- miEAA web server tutorial
- Browsable Web API Tutorial
- Command Line Interface and Python/R API Documentation
MiEAA browsable API tutorial
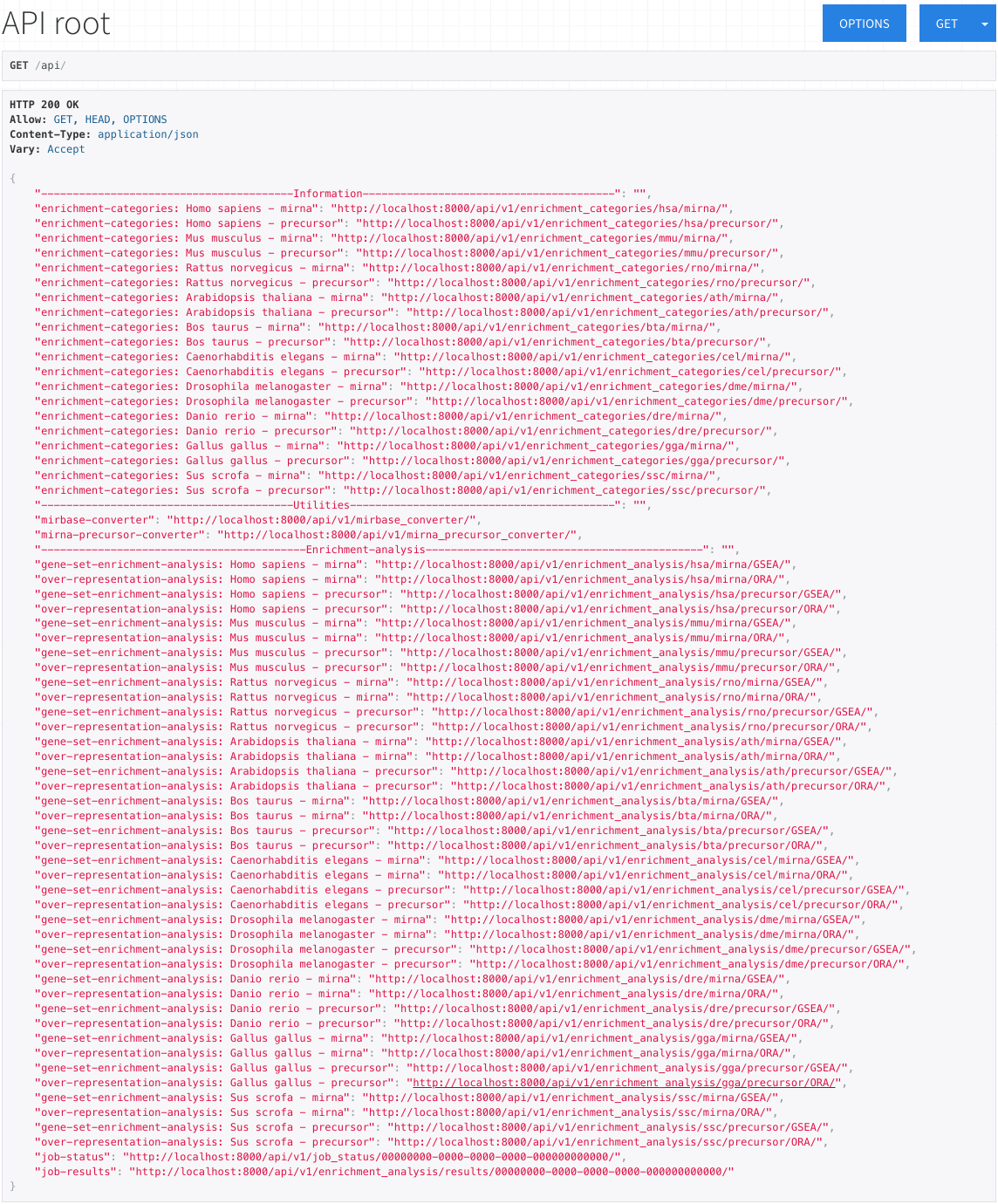
API root
The API root displays all publicly available miEAA endpoints.
From this page, you can access specific enrichment analyses and converters, or view the possible enrichment categories for each species.
The URLs corresponding to each option can be directly clicked or copy-pasted into your web browser.
The root view is divided into "Information", "Utilities", and "Enrichment Analysis" in order to easily access the desired functionality.
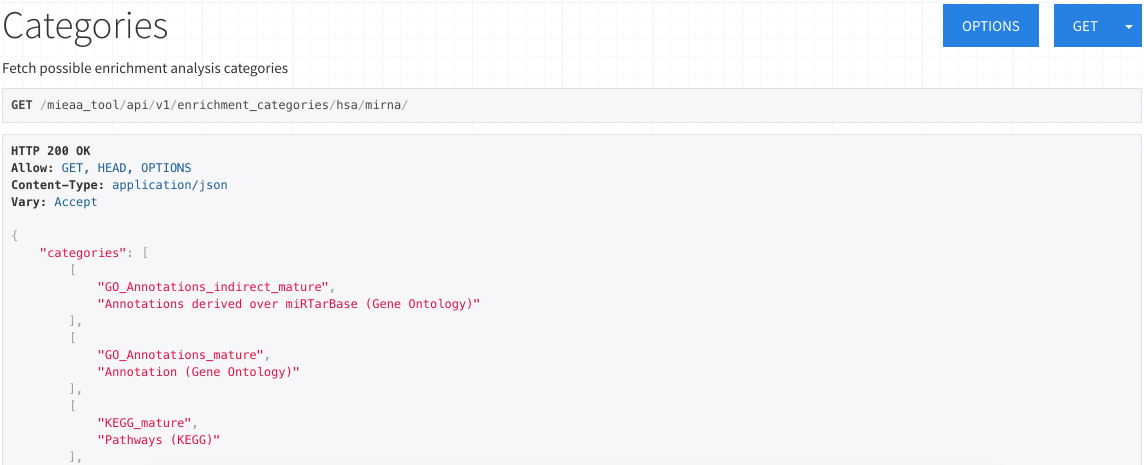
Information
The information heading contains the URLs to the enrichment categories available for each species and miRNA/precursor.

After selecting the desired URL, you will be able to view all the sources available for that input.
The first value in each item corresponds to the GMT filename (without the extension), and the second value to it's description.
The categories can also be viewed and saved in json format by selecting it from the GET dropdown.
Utilities
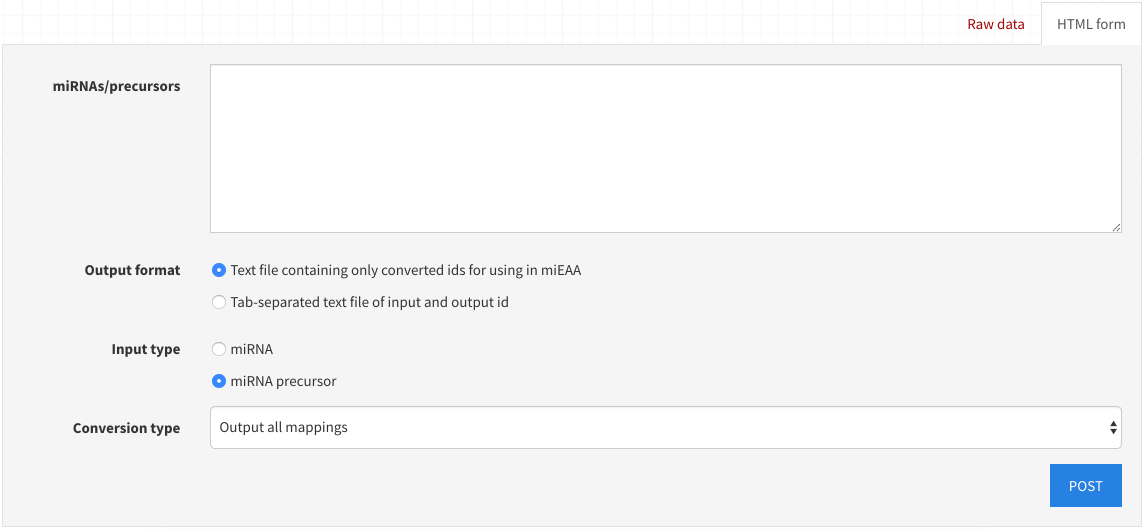
The utilities heading contains both the miRBase version converter and the precursor <-> miRNA converter.
![]()
For conversion, simply navigate to the desired URL, fill out the HTML form, and select POST.
You will be prompted to select a save file location.
Note that you can make a POST request containing a valid json payload from any fully-qualified remote client to the same endpoint(s).
Enrichment Analysis
Under the enrichment analysis heading, you can find the the links to GSEA and ORA analyses.
You can run an analysis on all supported species, for either miRNAs or their precursors.
The "job-status" and "job-results" URLs are only available after starting or completing an analysis, respectively.
These two job related endpoint URLs must contain the corresponding job's ID.

To start an analysis, follow any link with the desired combination of algorithm, input type, and species and fill in the resulting form.
If you need help understanding the fields, please refer to the miEAA web server tutorial.
To select multiple categories at once, hold down your keyboard's "control" key ("command" on Mac) and click on the available options.
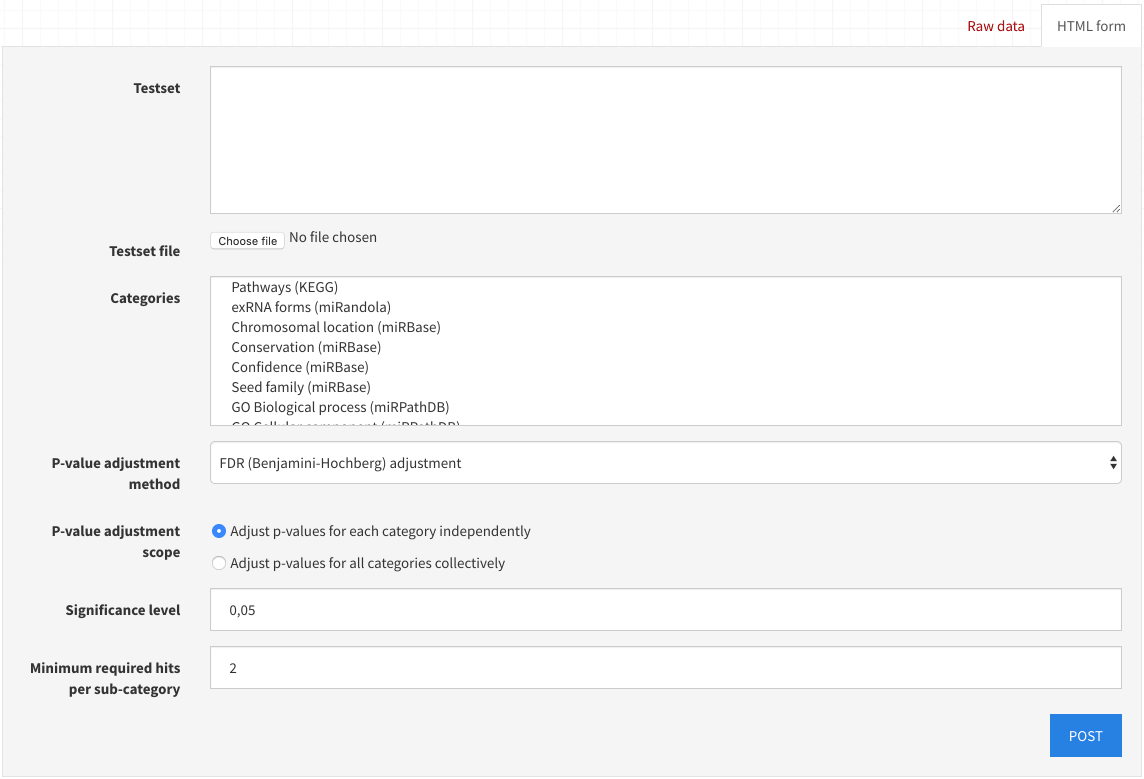
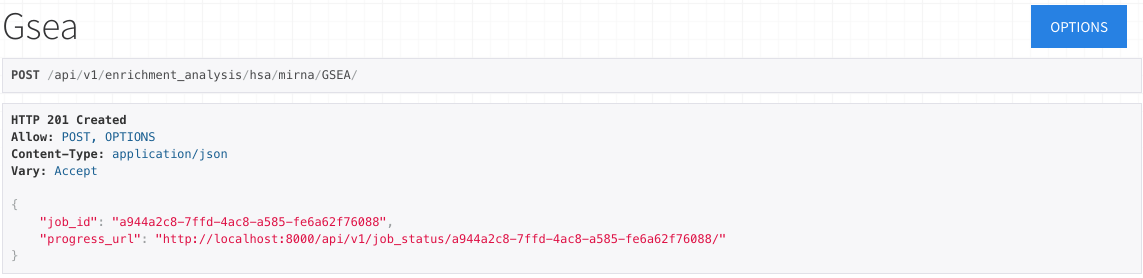
Upon selecting POST, the form will be submitted.
If the submission is valid, the response at the top of the page will display a job ID and progress URL.
An invalid submission will display a descriptive error message instead.
By selecting this progress URL, you may view the corresponding job's status.
Job Status
On the job status page, you may view the current progress of the running job.
Refreshing this page will update the status with the current percentage of completion.
Once a job is complete, results will be available and can be accessed via a newly transmitted results URL.
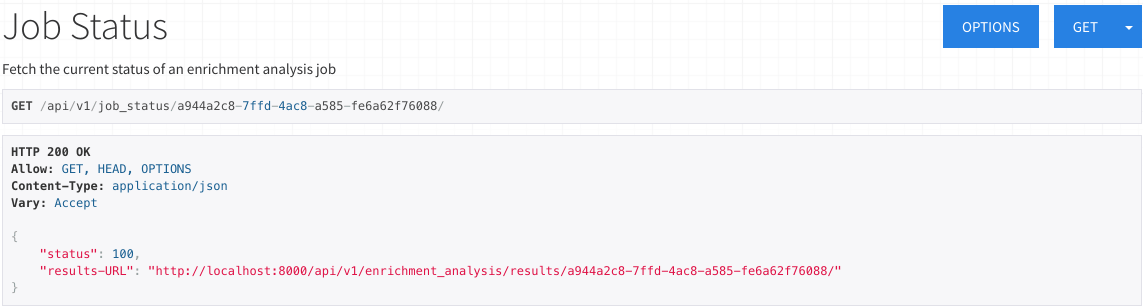
Please note that you may have to manually refresh this page to view the results URL.
In a custom remote-client you may call the same URL repeatedly to update the current status, however, please note that
issuing too many requests per second will temporarily close the connection to prevent any exploits and to improve stability of our service.
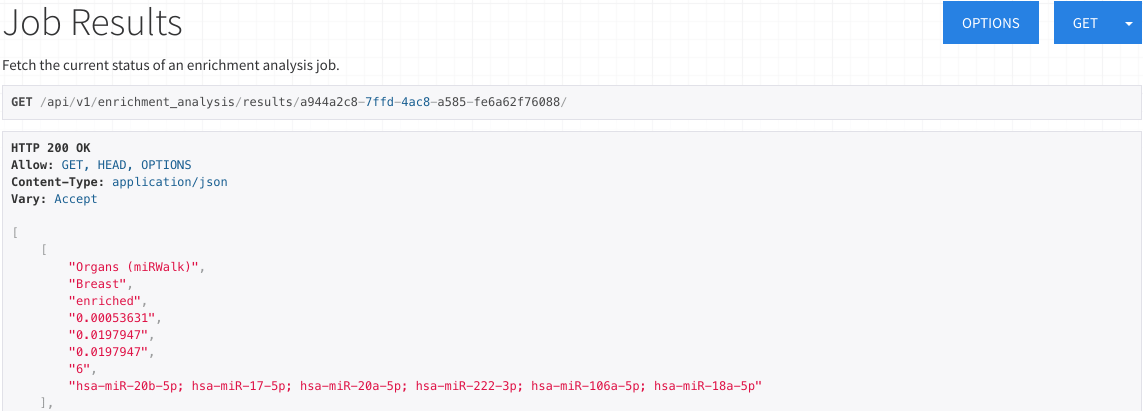
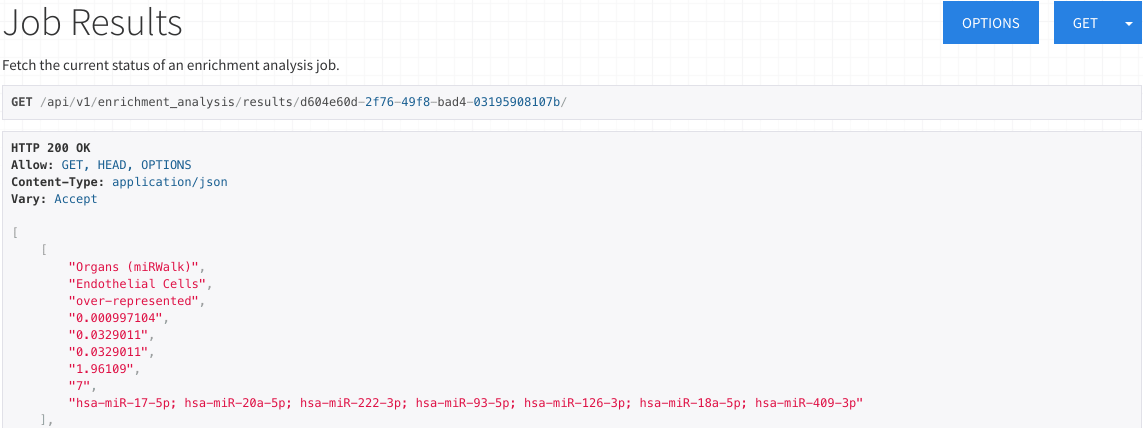
Results for ORA
The ORA results display a list all significantly under-represented or over-represented subcategories.
Each item contains the category, subcategory, whether the subcategory was over-represented or under-represented,
p-value, adjusted p-value, q-value (Benjamini-Hochberg FDR adjustment), expected count, observed count,
and miRNAs/precursors of the testset contained in the subcategory.
Results can also be viewed or saved in json and csv formats.
Please note that for very large result lists, the
performance of the website to display the results depends on your computer and browser and thus may degrade. We suggest to switch to a
client-based usage through python / R in such cases.
Results for GSEA
The GSEA results display a list of all significantly enriched or depleted subcategories.
Each item contains the category, subcategory, whether the subcategory was enriched or depleted,
p-value, adjusted p-value, q-value (Benjamini-Hochberg FDR adjustment), observed count,
and miRNAs/precursors of the testset contained in the subcategory.
Results can also be viewed or saved in json and csv formats.